Widefield (WF) and Confocal (CF) Microscopy might be sufficient to study cellular and tissue morphology and dynamics but are often not enough to accurately answer all biological questions. As a matter of fact, in WF and CF microscopy subcellular structures still remain inaccessible and hidden by the diffraction limit. Getting a further level of information could allow new discoveries and, in this regard, Super-Resolution (SR) Microscopy has become an increasingly popular and robust tool across life sciences to study fine cellular structures and sub-cellular components, as well as their dynamic processes. These types of experiments typically require overcoming the diffraction limit, doubling the lateral and axial resolution which can be achieved with WF microscopy. In the past few years there has been a parallel evolution of technology and applications, resulting in an exponential growth of published papers about SR techniques or showing SR in life science applications. However, despite the enormous potential of these techniques, there are still major limitations associated with SR, mostly related to its cost and complexity. Because of several negative connotations, including special sample preparation, poor depth penetration and prohibitive cost, SR is not yet routine in everyday laboratory life. For these reasons, in SIMTRUM we are working to make SR accessible to all researchers and to advance the potential of their scientific discoveries, moving them to a next level. Our newest product, the SIM Basic, was developed with this purpose and it is the first SR module based on structured illumination compatible with any existing upright or inverted microscope with a camera port, it is as easy to use as a CF microscope and it enables scientists to access deep data about their biological samples. In particular, based on a multi-spot structured illumination system, the SIM Basic is able to study cellular structures up to an XY resolution of ~100 nm and a super-resolved optical sectioning, with Z resolution up to ~300 nm, can be obtained using both high (60X-100X) and low (20X-40X) magnification objectives. The SIM Basic is designed to work with samples of thickness comparable to those used in CF microscopy and in this Application Note we are going to focus on super-resolved deep imaging about different neurobiological samples at high magnification.
|
| Super-Resolution imaging of cleared samples at low magnification |
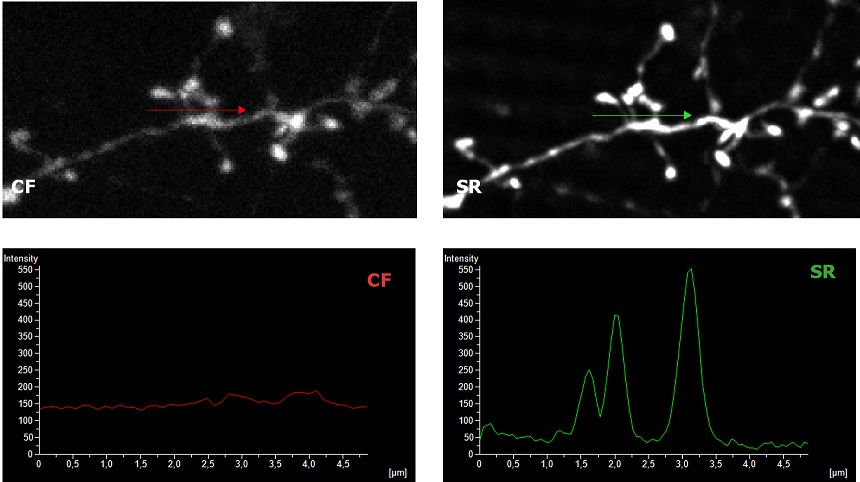
| To demonstrate the SIM Basic optimal performance also with low magnification objectives, we compared Widefield (WF), Confocal (CF), and SR images in complex and thick cleared biological samples using a CFI Plan Apo Lambda 20X air objective (NA 0.75, WD 1). In Figure 1, different visualizations of a cleared mouse intestine section (0.55 mm thickness) with blood vessels in green and nuclei in red are shown. In particular, thanks to the fast switch between three microscopy methods, we report a global comparison of WF, CF spinning disk and SR acquisitions of a volume of 125 um (Figure 1A) and a 3D movie of the SIM Basic SR acquisition showing fine details of intestinal villi (Figure 1B). In Figure 2, a cleared mouse brain section (0.55 mm thickness) with GFP-expressing neurons is shown. Notably, we acquired a 150 um Z stack of an intricate neuronal network shown as maximum intensity projection (MIP) (Figure 2A) and 3D volume view (Figure 2B). As nicely appreciated from these images, switching from WF, CF and SR modalities allows reaching an increasing resolution gain that enhances the fine details of the neuronal network even at high thickness. Moreover, we decide to compare the 20X SR acquisition with a 60X CF acquisition and, as demonstrated in figure 2C and 2D, the acquisitions with the SIM Basic not only allow to have an evident resolution improvement compared to the 20X CF acquisitions, but it enables to reach an image quality comparable to those obtained with the CF spinning disk equipped with a CFI Plan Apo Lambda 60X oil objective (NA 1.4, WD 0.13). In addition to a noticeable enhancement in neuronal details quality with respect to a 20X CF (Figure 2D), the use of the SIM Basic with 20X objective allows going even deeper into the sample than what is permitted by the maximum working distance of a common 60X oil objective (130 um) (Figure 2C).
|
A 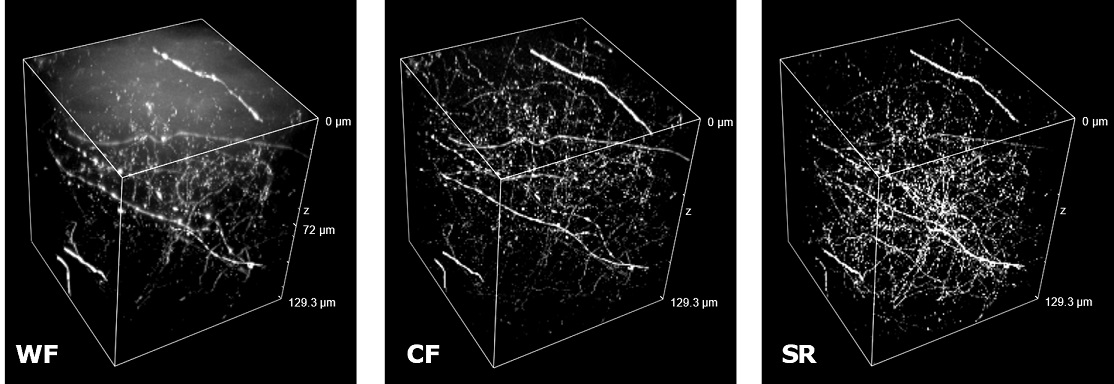
|
B 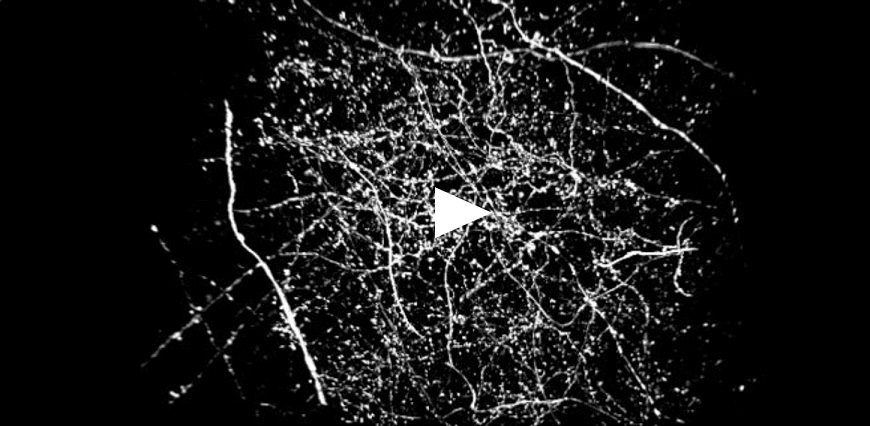
|
C
|
Figure 1: (A) Comparison of WF, CF spinning disk and SIM Basic SR 3D volume views. (B) 3D movie of SIM Basic SR acquisition, 130 um thickness. (C) Visualization and intensity profile comparison of dendritic spines acquired with SIMTRUM SpinDisk Advance CF spinning disk and SIM Basic SR system. |
These images demonstrate that, thanks to a super-resolved optical sectioning with Z resolution up to ~ 300 nm, the SIM Basic allows the addition of a deeper level of detail to large CF acquisitions. The SIM Basic is in fact designed to work with samples of thicknesses comparable to those used with CF microscopy, giving super-resolved data over 50 µm Z depth. This means that the only limitation is given by the working distance of the objective and that more meaningful data can be obtained from native heterogeneous complex samples. |
|
| Super-Resolution imaging of conventionally prepared samples |
| In uncleared samples, the different refractive index of the main biological components, such as proteins, lipids and water, causes a greater scatter of light when it passes through the tissue. As a result, light does not easily pass deep into the sample causing problems in deep SR imaging. With SIM Basic, we are able to offer SR also inside routinely prepared thick samples, without the need of special sample preparation, and we can super-resolve 3D samples of a size comparable to what is routinely imaged with a CF microscope. For this purpose, we tested several scattering neurobiological samples and in Figure 2 different visualizations of conventionally prepared mouse brain sample (100 um thickness) with GFP-expressing neurons are shown. In particular, we focus on dendritic spines and, as nicely reported in Figure 2A and 2B, these detailed protrusions are much more detectable and clearer in the super- resolved images compared to WF and CF spinning disk data. Moreover, the super-resolved 3D rendering of this sample (Figure 2C) demonstrates that it is possible to appreciate this nanometer-sized protrusion even 60 um deep inside the sample. |
|
A 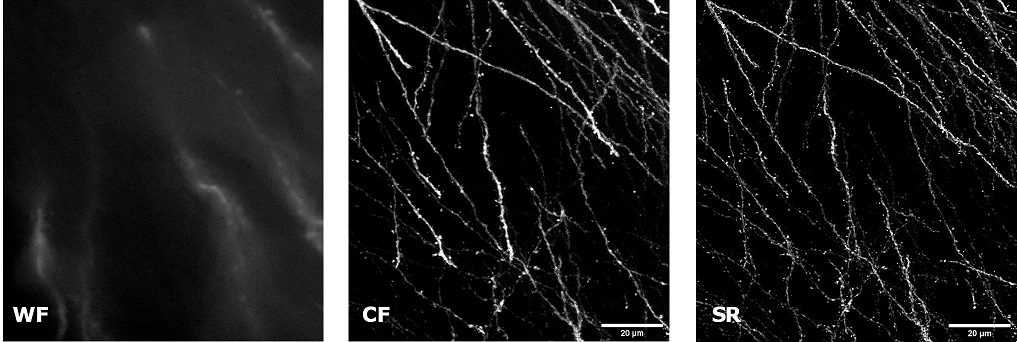
|
B 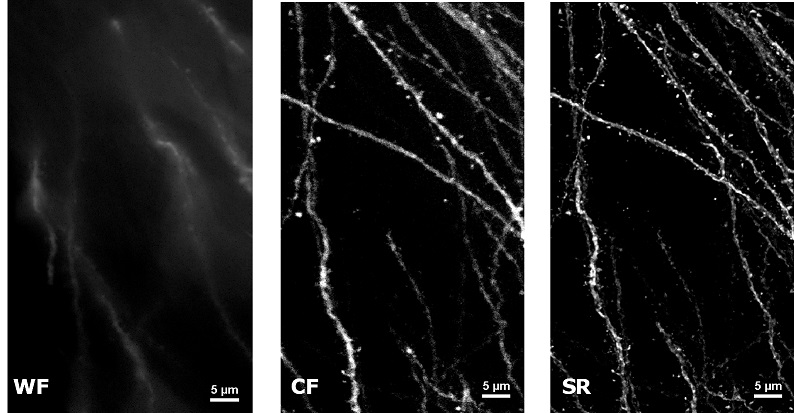
|
C 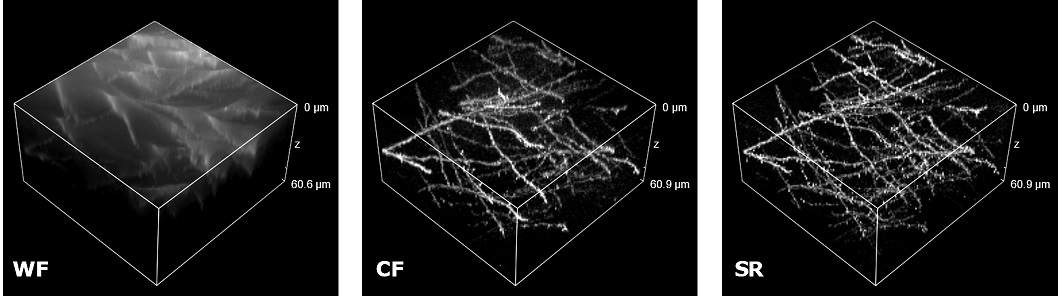
|
| Figure 2: Comparison of WF, CF spinning disk and SIM Basic SR acquisitions of conventionally prepared mouse brain sample with GFP-expressing neurons. (A) Maximum intensity projection from Z stack, (B) neuronal structures details and (C) 3D volume views, 60 um thickness. These images were acquired with SIMTRUM SpinDisk Advance CF spinning disk system coupled with SIM Basic SR add on
|
| Figure 3 represents different visualization of a mouse brain (250 um thickness) with GFP-expressing microglia and, despite the presence of biological structures that inevitably create a scattering of light in an uncleared tissue, it was possible to perform acquisitions in depths comparable to those normally reached by a CF microscope, obtaining a notable increase in resolution and image quality (Figure 3A). This is nicely shown in Figure 3B and 3C where we focused on a particular microglial cell. The morphological features were observed and, thanks to the easy and fast switching between WF, CF spinning disk and SR, it was possible to enhance fine cellular structures otherwise difficult to appreciate, proposing the SIM Basic as a powerful tool for cell morphology studies. The use of SIM Basic is not restricted to fixed specimens only, but thanks to a temporal resolution of > 10fps (in a field of view of 1024x1024 pixels), and minimizing light exposure and therefore the risks of photo-toxicity, the SIM Basic high-speed acquisitions allow applying SR microscopy also for in vivo studies.
|
A 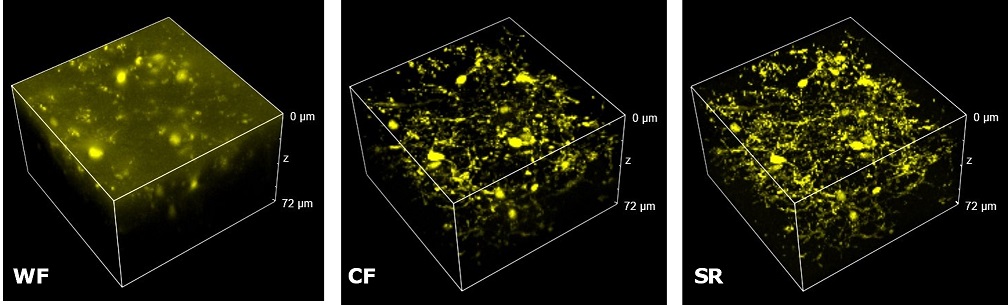
|
B 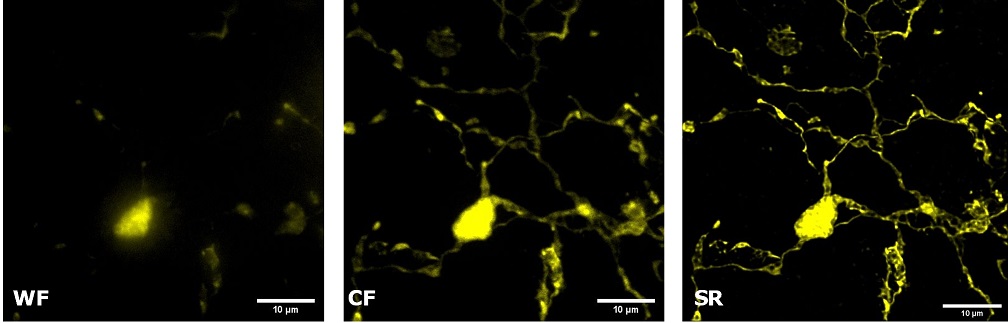
|
C 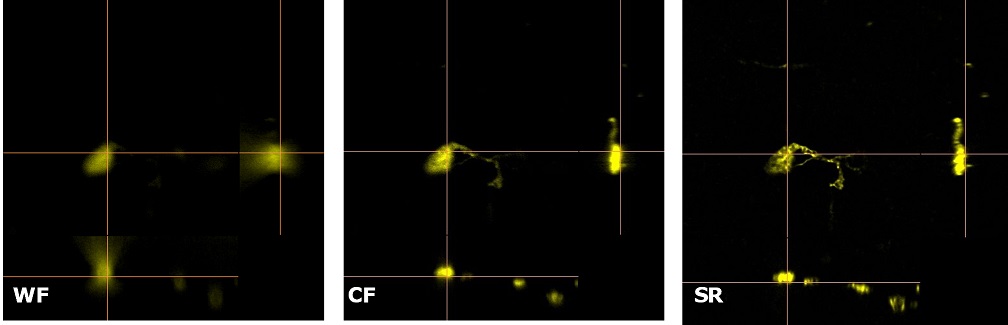
|
| Figure 3: Comparison of WF, CF spinning disk and SIM Basic SR acquisitions of conventionally prepared mouse brain sample with GFP-expressing microglia. (A) 3D volume views, 72 um thickness, (B) maximum intensity projection and (C) orthogonal views showing single microglial cell details. These images were acquired with SIMTRUM SpinDisk Advance CF spinning disk system coupled with SIM Basic SR add on. |
|
| In Figure 4 we report a comparison of CF spinning disk and SR acquisitions of a live transgenic Caenorhabditis elegans expressing a genetically encoded fluorescent Ca2+ indicator (green) and a pan-neuronal marker (red). The transgenic nematode was confined in a microfluidic device and paralyzed with 1 mM levamisole, an acetylcholine receptor-specific agonist. This simple immobilization, together with the SIM Basic high-speed temporal resolution, was sufficient for making super- resolution microscopy possible also on live samples. In particular, we report 3D volume reconstructions (Figure 4A) to then focus on head sensory neurons and interneurons (Figure 4B) obtaining a remarkable increase in resolution and image quality with respect to CF acquisitions. All together, these data demonstrate that the SIM Basic is able to work with samples of thickness comparable to those normally used in CF microscopy, giving super-resolved data over 50 um Z depth. This means that more meaningful data can be obtained from native heterogeneous complex samples using routine preparation protocols.
|
A 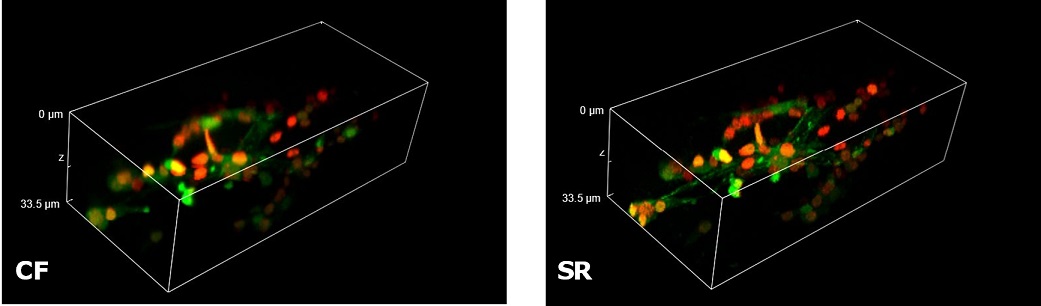
|
B 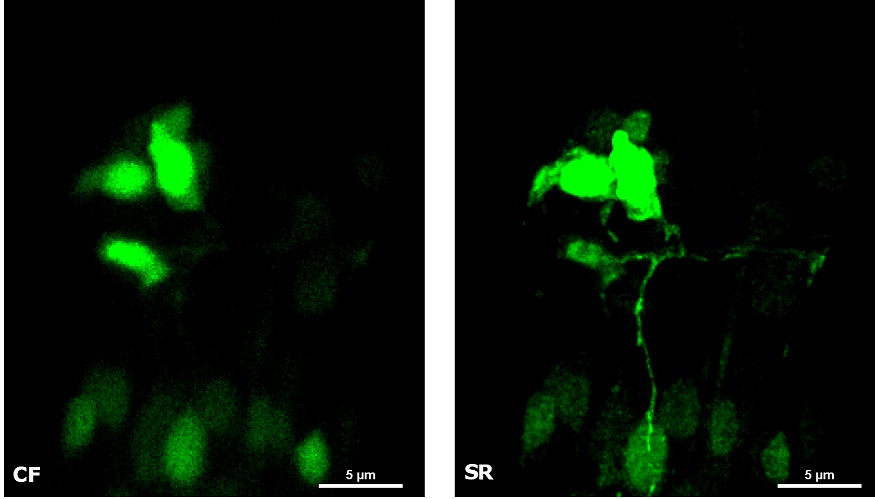
|
Figure 4: CF and SIM Basic SR acquisitions of a live transgenic Caenorhabditis elegans expressing a genetically encoded fluorescent Ca2+ indicator (green) and a pan-neuronal marker (red). (A) 3D volume visualizations, 33.5 um thickness, and (B) maximum intensity projection from Z stack showing head neurons details. These images were acquired with SIMTRUM SpinDisk Advance CF spinning disk system coupled with SIM Basic SR add on. |
|
| From single channel to fast multi-channel imaging at high resolution |
Multi-channel imaging is often used for the study of the interactions between different cell types within the tissue. For this purpose, we have designed the SIM Basic to work with the full spectrum from 400 to 750 nm in order to offer the max flexibility in fluorophores choice and the optimal multichannel imaging without spectral overlap. In Figure 5 a mouse brain tissue section showing neurons with dendritic spines (yellow), microglia (magenta), astrocytes (white) and DNA (cyan) is shown, in a total volume of 30 um. These images demonstrate that with SIM Basic it is possible not only to easily switch from WF to CF and SR microscopy technique, but also that fast and deep multi- color imaging of conventionally stained sample is possible with SR too. Moreover, taking advantage of the specifications of our CF spinning disk system, we can also do more and use the dual-camera function to simultaneously acquire multiple channels, speeding up the acquisitions. In conclusion, all these data taken together demonstrate the plasticity of the system. Combining the SIM Basic with a CF microscopy allows having the flexibility to choose a region of interest and easily switch to SR for revealing the desired part in subcellular details, doubling resolution with just one click. We believe that SR should be accessible for all scientists to progress their research. Biology happens in 3D and it is essential to explore all the three dimensions in SR to expand the discovery potential.
|
A 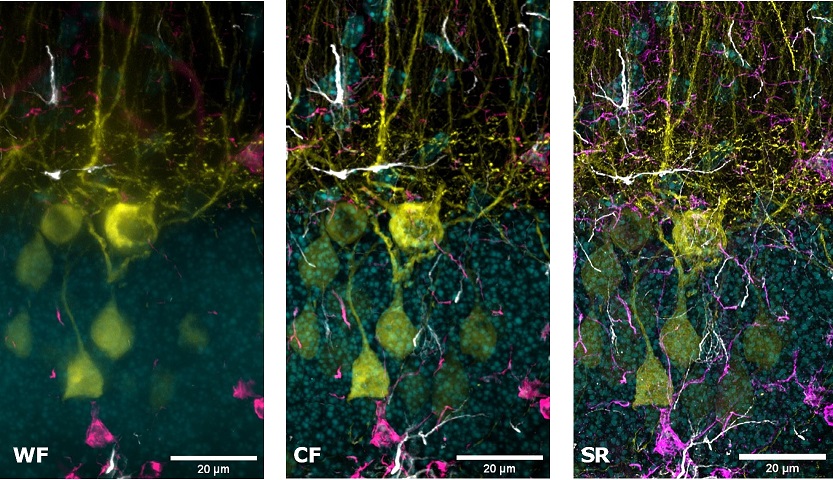
|
B
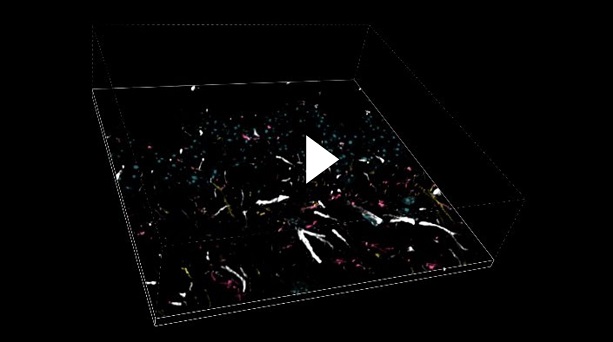
|
Figure 5: WF, CF spinning disk and SIM Basic SR acquisitions of conventionally prepared mouse brain tissue section showing neurons with dendritic spines (yellow), microglia (magenta), astrocytes (white) and DNA (cyan). (A) Maximum intensity projection from Z stack and (B) 3D movie of SIM Basic SR acquisition, 30 um thickness. These images were acquired with SIMTRUM SpinDisk Advance CF spinning disk system coupled with SIM Basic SR add on.
|
| Microscopy Methords |
All the acquisitions of this Application Note were performed through a Nikon Eclipse Ti2 microscope equipped with SIMTRUM SpinDisk Advance spinning disk system coupled with SIM Basic super-resolution add on, LDI laser illuminator (89 North) and Prime BSI Scientific CMOS (sCMOS) camera with 6.5 um pixels (Photometrics). We used a CFI Plan Apo Lambda 60x oil objective (NA 1.4, WD 0.13) and performed Z-stack acquisitions with 0.3 um Z step size. Mouse Brain samples shown in Figure 2 were kindly provided by Dr Alvaro Crevenna and Dr Emerald Perlas , EMBL -Rome. Mouse Brain samples represented in Figure 3 were kindly provided by Prof Davide Ragozzino and Dr Laura Ferrucci Sapienza Università di Roma, Dept. Neuroscience . C.elegans shown in Figure 4 were kindly provided by Viola Folli and Dr Enrico Lanza Center for Life Nano- & Neuro-Science (CLN2S@Sapienza Università di Roma- Istituto Italiano di Tecnologia ). Mouse Brain sections represented in Figure 5 were kindly provided by Prof Silvia Di Angelantonio and Dr Federica Cordella Center for Life Nano- & Neuro-Science(CLN2S@Sapienza Università di Roma-Istituto Italiano di Tecnologia ).
|